About TopOMetry
TopOMetry is a high-level python library to explore data topology through manifold learning. It is compatible with scikit-learn, meaning most of its operators can be easily pipelined.
Its main idea is to approximate the Laplace-Beltrami Operator (LBO). This is done by learning properly weighted similarity graphs and their Laplacian and Diffusion operators. By definition, the eigenfunctions of these operators describe all underlying data topology in an orthonormal eigenbasis. These eigenbases are special versions of Diffusion Maps, Laplacian Eigenmaps or Kernel Eigenmaps. New topological operators are then learned from such eigenbasis and can be used for clustering and graph-layout optimization (visualization).
For more information, please see our pre-print.
TopOMetry is designed to handle large-scale data matrices containing extreme sample diversity, such as those generated from single-cell omics. It includes wrappers to deal with AnnData objects using scanpy.
Tutorials
If you haven’t already, install topometry and start using it (quick-start).
For a first introduction, check the tutorial with MNIST handwritten digits.
To explore TopOMetry for document embedding, see the tutorial using the Newsgroups dataset
To dive deeper into TopOMetry classes, see the classes tutorial.
To explore how TopOMetry estimate local and global intrinsic dimensionality, check the dimensionality estimation tutorial.
To learn how to systematically run and evaluate multiple models, check the evaluation tutorial
To see how TopOMetry can be integrated into different single-cell workflows, check the RNA velocity tutorial using scVelo
To use TopOMetry to embedd to non-euclidean spaces (similarly to UMAP), see the tutorial on non-Euclidean spaces.
All of the tutorials are also freely available as Jupyter Notebooks in this separate repository.
When it is best to use TopOMetry
This is a frequently asked question with a simple answer: always, unless the data tells you so.
One should never assume a priori that a method will be the best with every single dataset. TopOMetry allows users to explore high-dimensional data in several ways and to compute many different representations for it. It is not claimed to be superior to every other method a priori. Instead, it allows practitioners to see which method works best for their particular use case by themselves, based on quantitative and qualitative assessments.
In all tested cases so far, TopOMetry models outperformed the classical PCA-based approach used in single-cell genomics, and in most of them it has also outperformed stand-alone UMAP. However, that should never be considered true without looking at the data and how these models perform on it. The aim here is to provide users with plenty of options and allow them to pick which is the best for them, empowering them with evidence-based decisions.
When not to use TopOMetry
First and foremost, when you do not have enough samples to safely assume that the manifold hypothesis holds true. In addition, one should consider that TopOMetry does not currently support neither including new data without recomputing decompositions, nor inverse transforms. If that is critical to your production workflow, then TopOMetry is problably not be the best option, and you might prefer to use UMAP or topological autoencoers. However, even in that case, you should consider TopOMetry in some of your data to evaluate whether your current workflow is generating reliable embeddings, or to estimate its intrinsic dimensionalities in order to properly build an adequate architecture.
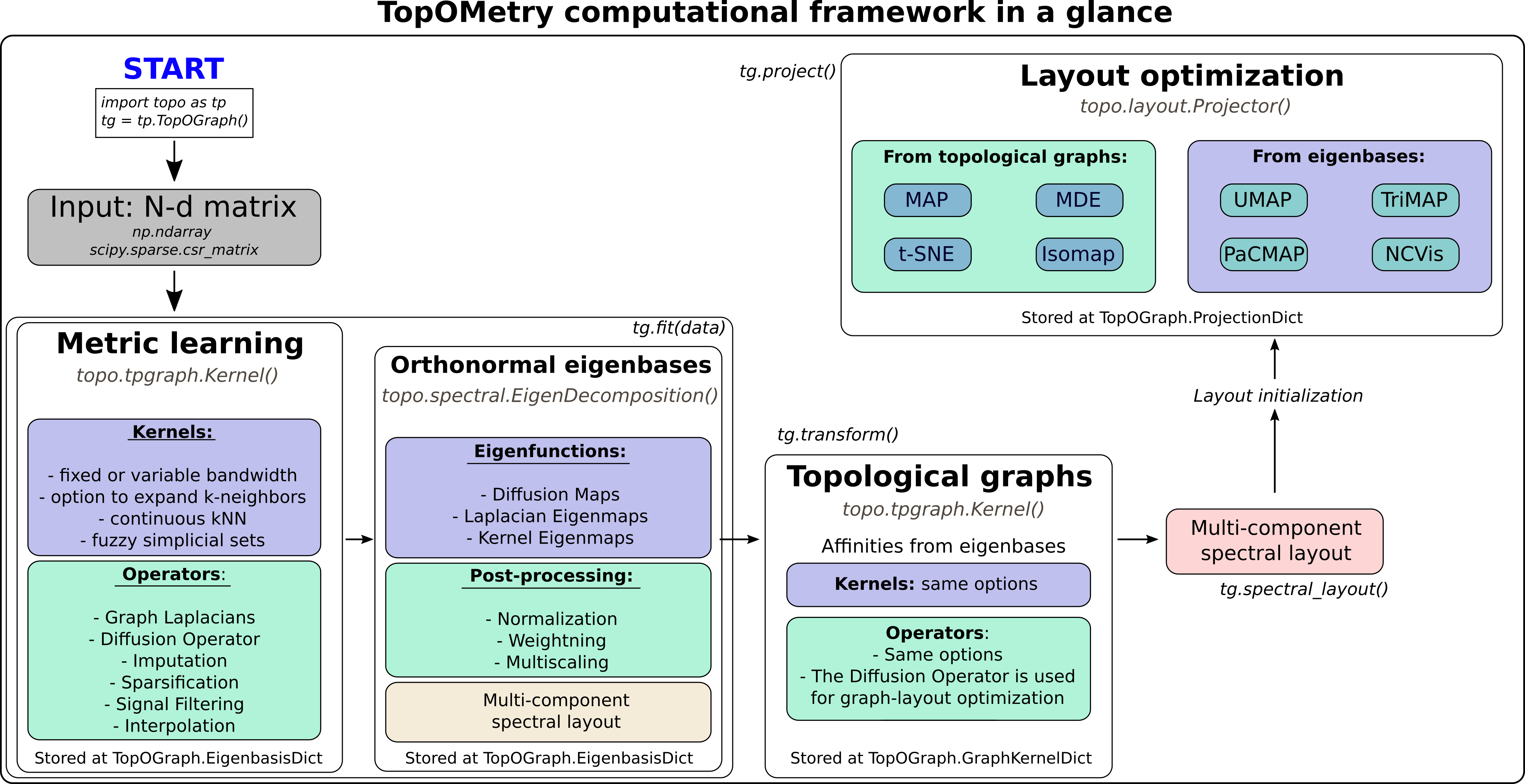
TopOMetry classes
TopOMetry is centered around four classes of scikit-learn-like transformers:
Kernel - learns similarities and builds topological operators that approximate the LBO.
EigenDecomposition - obtains and post-processes eigenfunctions.
Projector - handles graph-layout optimization methods.
TopOGraph - orchestrates analysis by employing the above estimators and others.
The following diagram represent how TopOMetry uses these transformers to learn topological operators, their eigenfunctions as orthonormal eigenbases, topological operators of these eigenfunctions and graph projections of these graphs or eigenbases:
Citation
@article {Sidarta-Oliveira2022.03.14.484134,
author = {Sidarta-Oliveira, Davi and Velloso, Licio A},
title = {A comprehensive dimensional reduction framework to learn single-cell phenotypic topology uncovers T cell diversity},
elocation-id = {2022.03.14.484134},
year = {2022},
doi = {10.1101/2022.03.14.484134},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2022/03/17/2022.03.14.484134},
eprint = {https://www.biorxiv.org/content/early/2022/03/17/2022.03.14.484134.full.pdf},
journal = {bioRxiv}
}



